Deep Learning – Natural Language Processing
OPSA : Order Preserving token Shuffling Augmentation with wargame simulation dataset
Objective
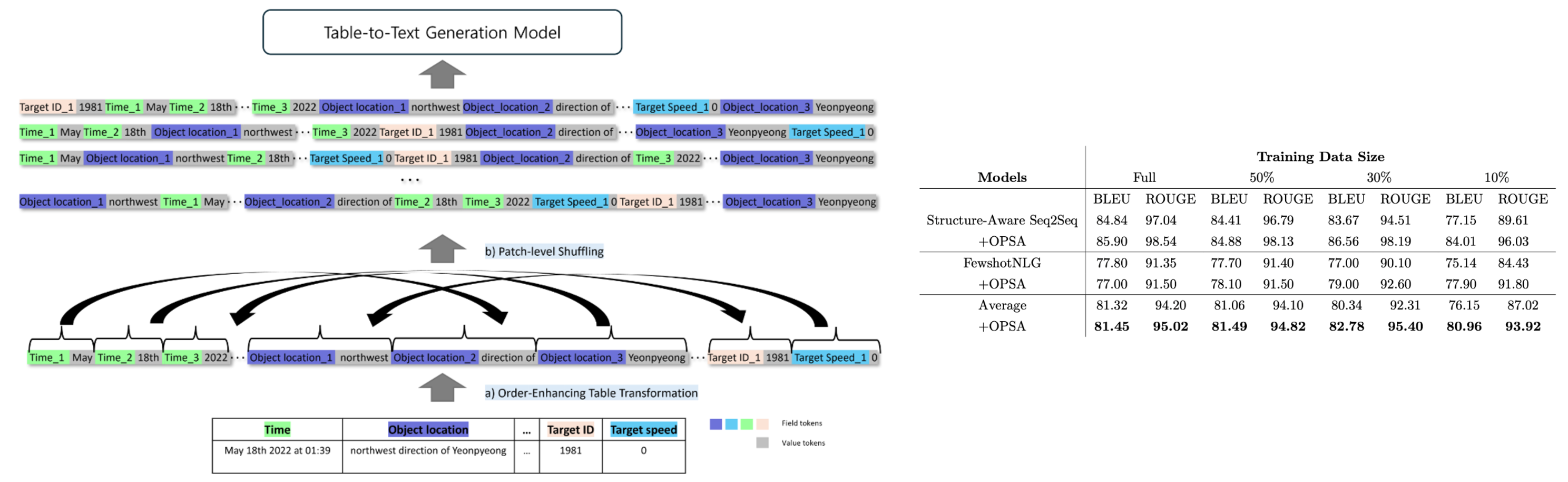
Table-to-text generation is the task of producing textual descriptions from structured tabular data while addressing challenges such as limited domain-specific datasets and the risk of generating hallucinated content. Existing methods do not simultaneously mitigate data scarcity and ensure accuracy without relying on extensive additional neural network training. In this study, we propose the Order Preserving Token Shuffling Augmentation (OPSA), a novel data augmentation methodology that shuffles tokens within a table while preserving their overall order, thereby improving the quality, diversity, and contextual relevance of generated text. By reducing the likelihood of hallucination and enhancing domain-specific outcomes—particularly in military simulations—OPSA provides a cost-effective solution for more effective information dissemination in specialized domains.
Data
This study utilized battle scenario data generated through military operation simulations using the Changjo21, Changgong, and Cheonghae models from South Korea, with actual names and location information modified for security reasons.
Related Work
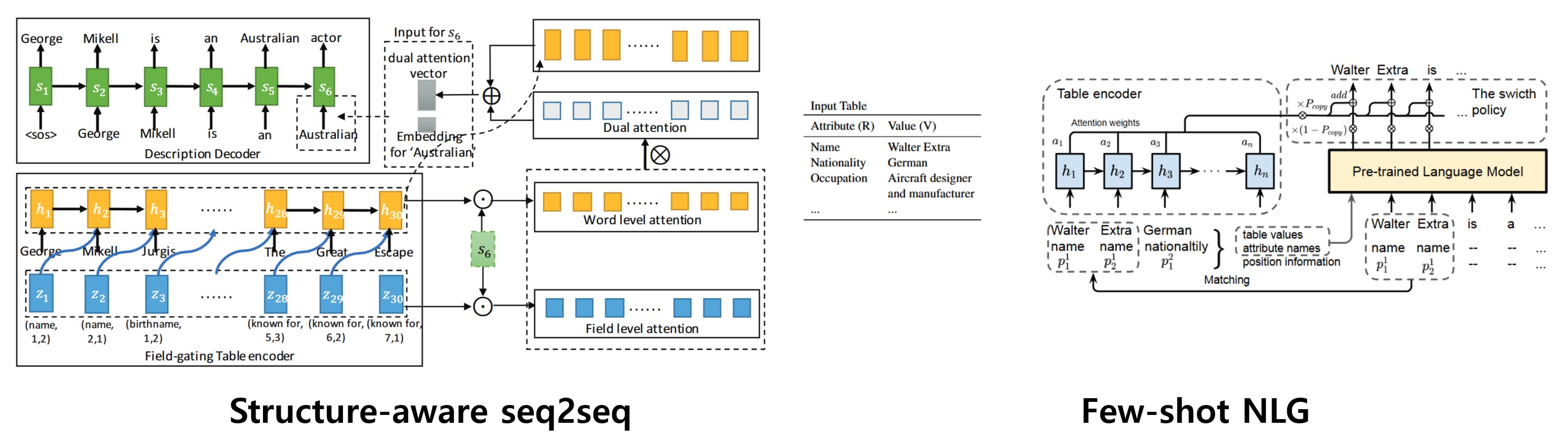
Structure-aware seq2seq[1]: Structure-aware seq2seq is a framework designed for data-to-text generation that incorporates structured information from the input data, allowing the model to better understand and represent the hierarchical relationships in the data, thus enhancing the coherence and relevance of the generated text.
Few-shot NLG[2]: Structure-aware seq2seq is a framework designed for data-to-text generation that incorporates structured information from the input data, allowing the model to better understand and represent the hierarchical relationships in the data, thus enhancing the coherence and relevance of the generated text.
[2]Chen, Zhiyu, et al. “Few-shot NLG with pre-trained language model.” arXiv preprint arXiv:1904.09521 (2019).
Proposed Method
Order Preserving Token Shuffling Augmentation (OPSA) is designed to enhance table-to-text generation by augmenting training data without requiring additional neural network training. OPSA comprises two main components: the Order-Enhancing Table Transformation (OET) module and the patch-level token shuffling module. The OET module converts structured table data into a sequential format that integrates positional information, enabling more effective modeling of tabular structure. Meanwhile, the patch-level token shuffling module rearranges tokens while preserving their original order, thus maintaining the semantic integrity of the content. By retaining the sequence of patches corresponding to the same field, OPSA effectively augments the dataset and improves model performance when generating natural language descriptions from tabular data.
Similarity-Based Unsupervised Spelling Correction Using BioWordVec: Development and Usability Study of Bacterial Culture and Antimicrobial Susceptibility Reports
Objective
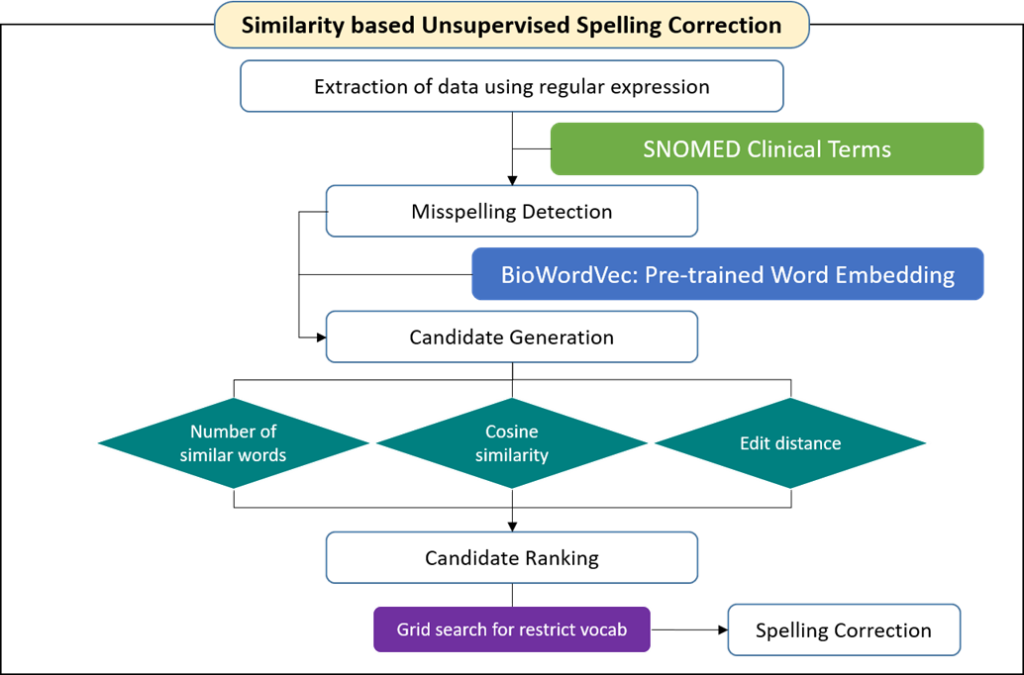
Spelling correction; natural language processing : In this research, we proposed a similarity-based spelling correction algorithm using pretrained word embedding with the BioWordVec technique. This method uses a character-level N-grams–based distributed representation through unsupervised learning rather than the existing rule-based method. In other words, we propose a framework that detects and corrects typographical errors when a dictionary is not in place.
Data
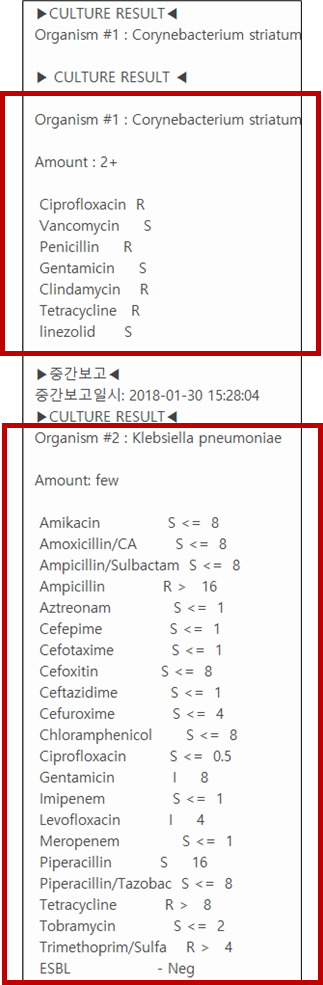
In this study, the bacterial culture and antimicrobial susceptibility reports from Korea University Anam Hospital, Korea University Guro Hospital, and Korea University Ansan Hospital were used. The bacterial culture and antimicrobial susceptibility report data were collected for 17 years (from 2002 to 2018), and in each year, reports for 1 month were used for the experiment. In total, 180,000 items were retrieved, with 27,544 having meaningful test results. Using the self-developed rule-based ETL algorithm, unstructured bacterial culture and antimicrobial susceptibility reports were converted into structured text data. After preprocessing through lexical processing, such as sentence segmentation, tokenization, and stemming using regular expressions, there were 320 types of bacterial identification words in the report. Among the extracted bacterial identification words, 16 types of spelling errors and 914 misspelled words were found.
Related Work
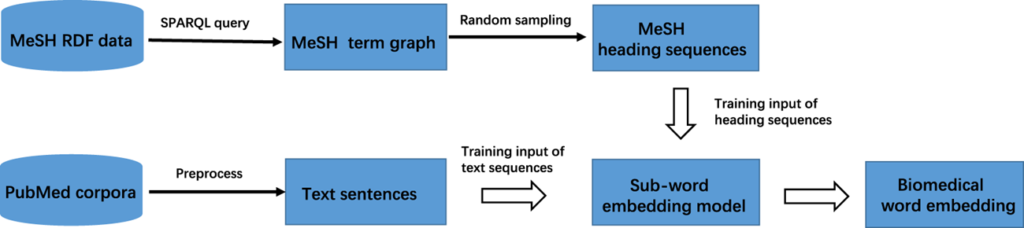
BioWordVec learns clinical record data from PubMed and MIMIC-III clinical databases using fastText. Based on 28,714,373 PubMed documents and 2,083,180 MIMIC-III clinical database documents, the entire corpus was built. The Medical Subject Headings (MeSH) term graph was organized to create a heading sequence and to carry out word embedding based on a sequence combining MeSH and PubMed. BioWordVec provided a 200-dimensional pretrained word embedding matrix.
Proposed Method
For detected typographical errors not mapped to Systematized Nomenclature of Medicine (SNOMED) clinical terms, a correction candidate group with high similarity considering the edit distance was generated using pretrained word embedding from the clinical database. From the embedding matrix in which the vocabulary is arranged in descending order according to frequency, a grid search was used to search for candidate groups of similar words. Thereafter, the correction candidate words were ranked in consideration of the frequency of the words, and the typographical errors were finally corrected according to the ranking.
AI consultation chatbot
Objective
Development of a consultation AI chatbot for providing telemedicine counseling solutions
Data
EMR dataset was used for training the chatbot model.
Related Work
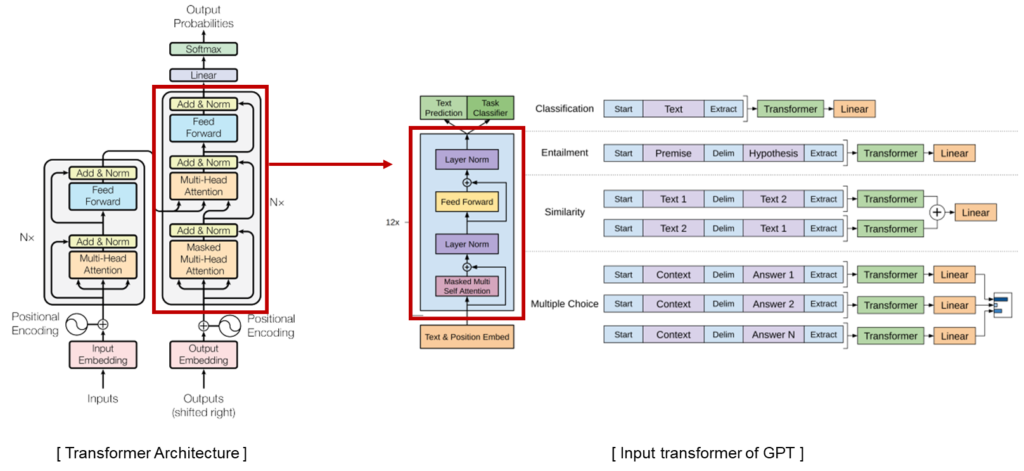
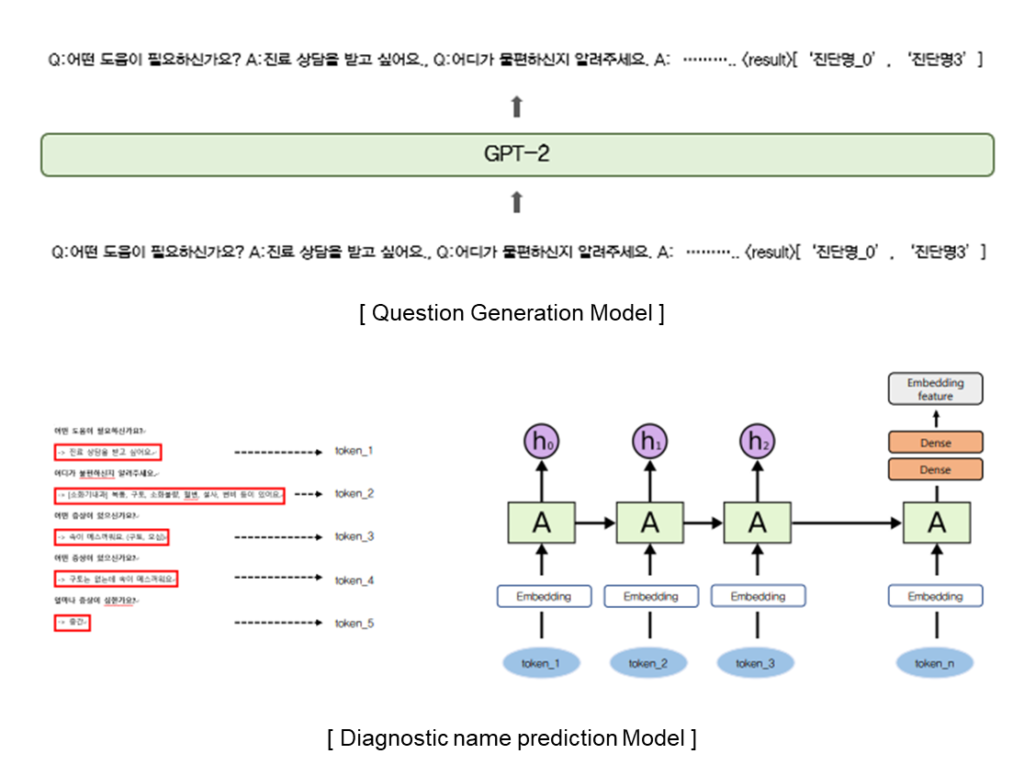
GPT-2 is a pretrained language model and optimized for sentence generation so that the next word in a given text can be well predicted. GPT-2 is a transformer decoder language model that has been learned with more than 40GB of text to overcome insufficient Korean performance.
Proposed Method
A Sequence consists of a list of questions and answers and a diagnostic name. It was used for fine-tuning GPT and predicting a diagnostic name. Therefore, it is possible to generate appropriate questions about the patient’s answers and finally predict the diagnostic name.
Research on Artificial Intelligence Writing Technology Using Natural Language Processing
Objective
Development of Novel Writing Platform based on “user input” through learning of various novel genres.
Data
KoGPT2 fine tuning is performed using novel text data. In the case of Semantic Role Labeling, we use ETRI Semantic Role Labeling Corpus for training SRL model.

Related Work
KoGPT2 is a pretrained language model and optimized for sentence generation so that the next word in a given text can be well predicted. KoGPT2 is a transformer decoder language model that has been learned with more than 40GB of text to overcome insufficient Korean performance.
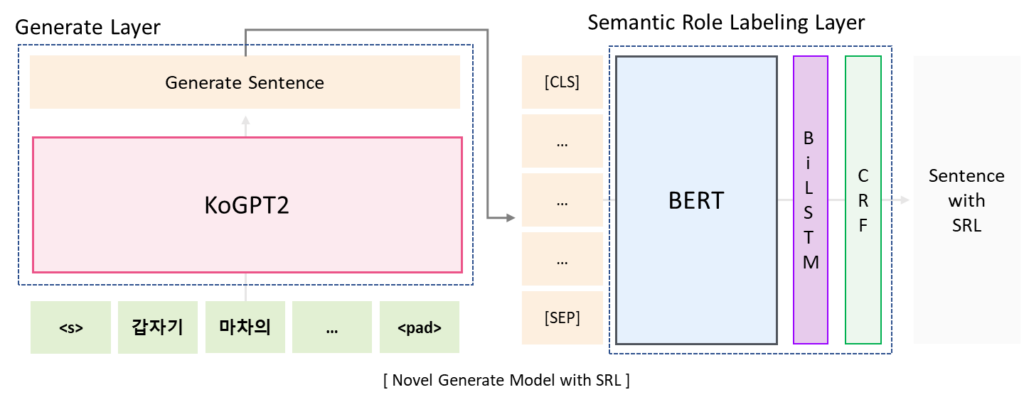
Proposed Method
It is a structure that combines Generate Layer for novel generation and SRL Layer for reflecting user input. When a sentence is entered, the following sentence is generated through KoGPT2 and the generated sentence is corrected through SRL layer.
Comparison of Data2Text models using Sequence-to-Sequence
Objective
1. Difficult to compare the performance of different Sequence-to-Sequence models.
- The data consists of a set of values and fields, and there are significant performance differences depending on how the model learns these structures.
- Therefore, we compare two representative models utilizing sequence-to-sequence under the same conditions to find a more effective methodology for learning the structure of the data.
2. There is a Problem that words that are not in the vocabulary, such as proper nouns, cannot be printed.
- Previous studies have proposed replacing “unk” token with attention distribution as a solution to the OOV(out-of-vocabulary) problem.
- However, this is a method that only applies if print out “unk” token.
Data
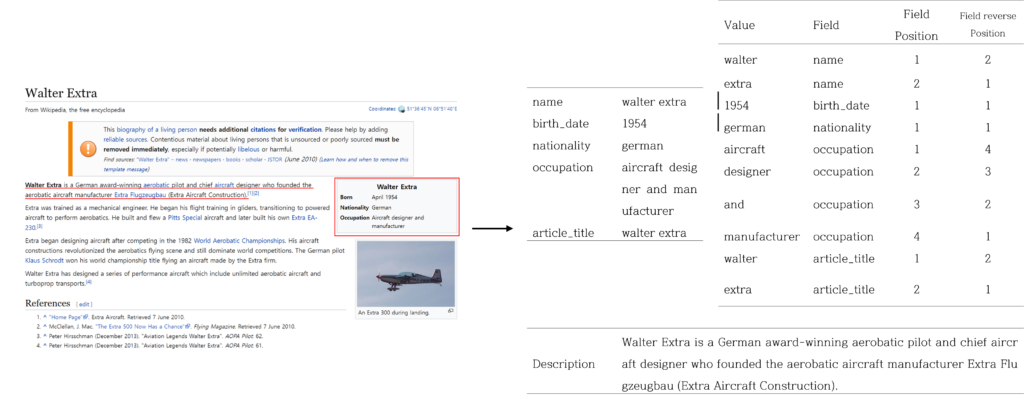
Wikibio Dataset : Wikibio consists of biographies of people recorded on Wikipedia. A given table is input data and the first sentence of the description is label.
Related Work
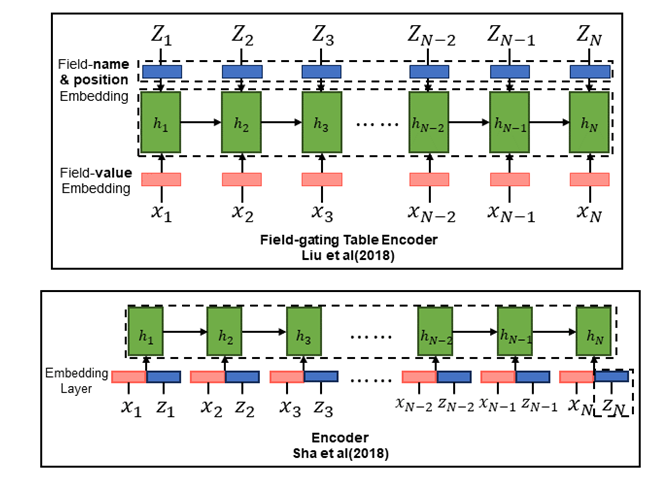
1. Table-to-text Generation by Structure-aware Seq2seq Learning
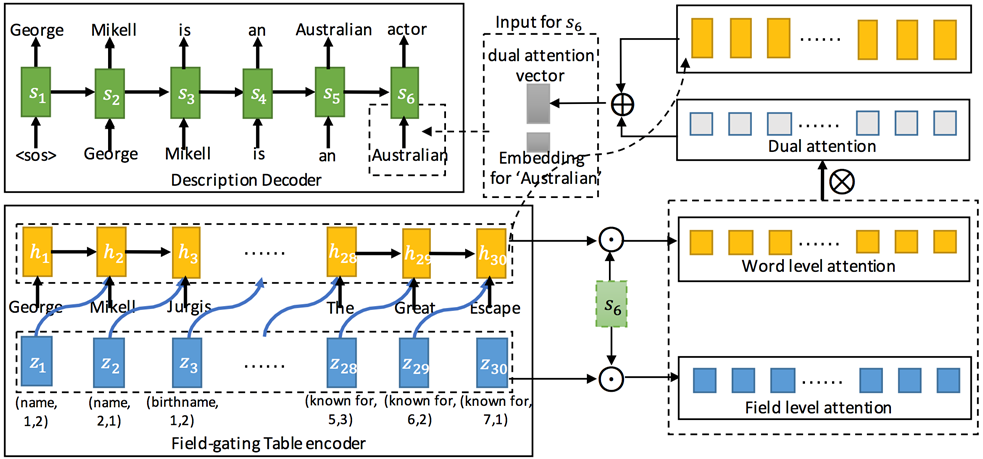
2. Order-Planning Neural Text Generation From Structured Data(Sha et al., 2018)
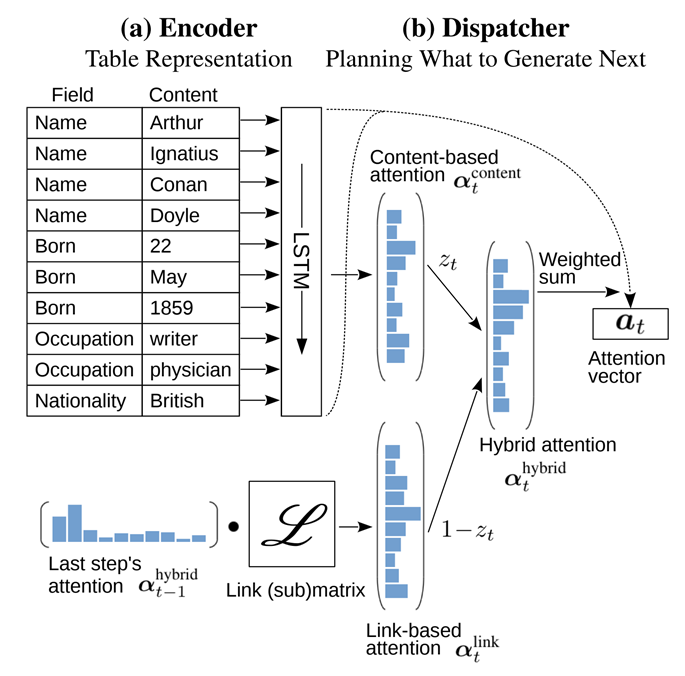
Proposed Method
We compare two representative models utilizing sequence-to-sequence under the same conditions to find a more effective methodology for learning the structure of the data. In addition, it adds a copy mechanism to improve performance by allowing the output of words that are not in the word vocabulary, such as proper nouns.
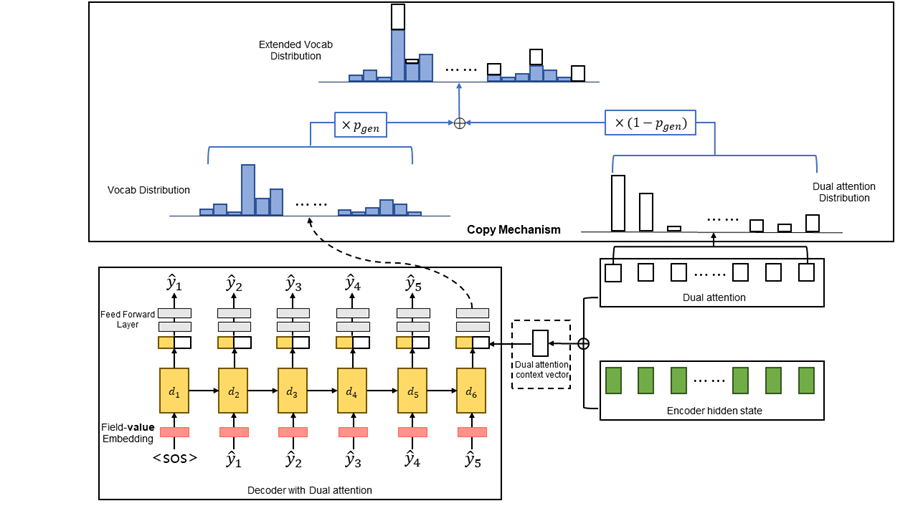
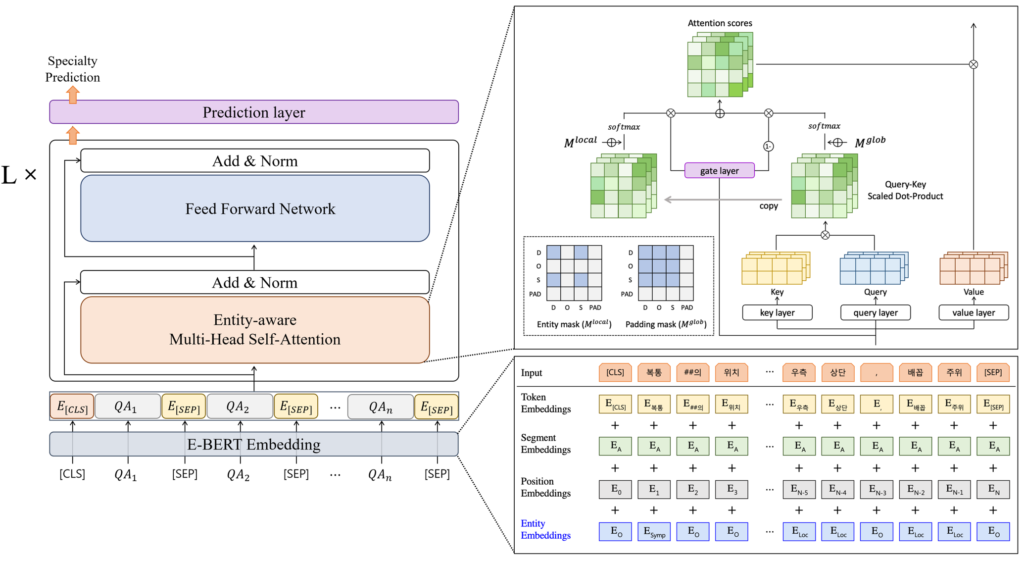
Entity-enhanced BERT for Medical Specialty Prediction Based on Clinical Questionnaire Data
Objective
- Medical text data include large amounts of information regarding patients, which increases the sequence length. Hence, a few studies have attempted to extract entities from the text as concise features and provide domain-specific knowledge for clinical text classification.
- However, It is still insufficient to inject entity information into the model effectively.
- We propose Entity-enhanced BERT (E-BERT), a single medical specialty prediction model by adding two modules that integrate entity information within BERT, to processes medical text.
Data
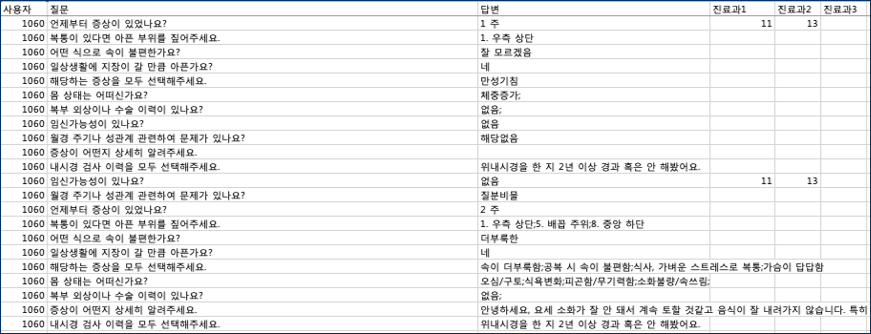
This is clinical questionnaire data containing information such as the patient’s symptoms, location of pain, disease, and lifestyle habits in a question and answer format.
Related Work
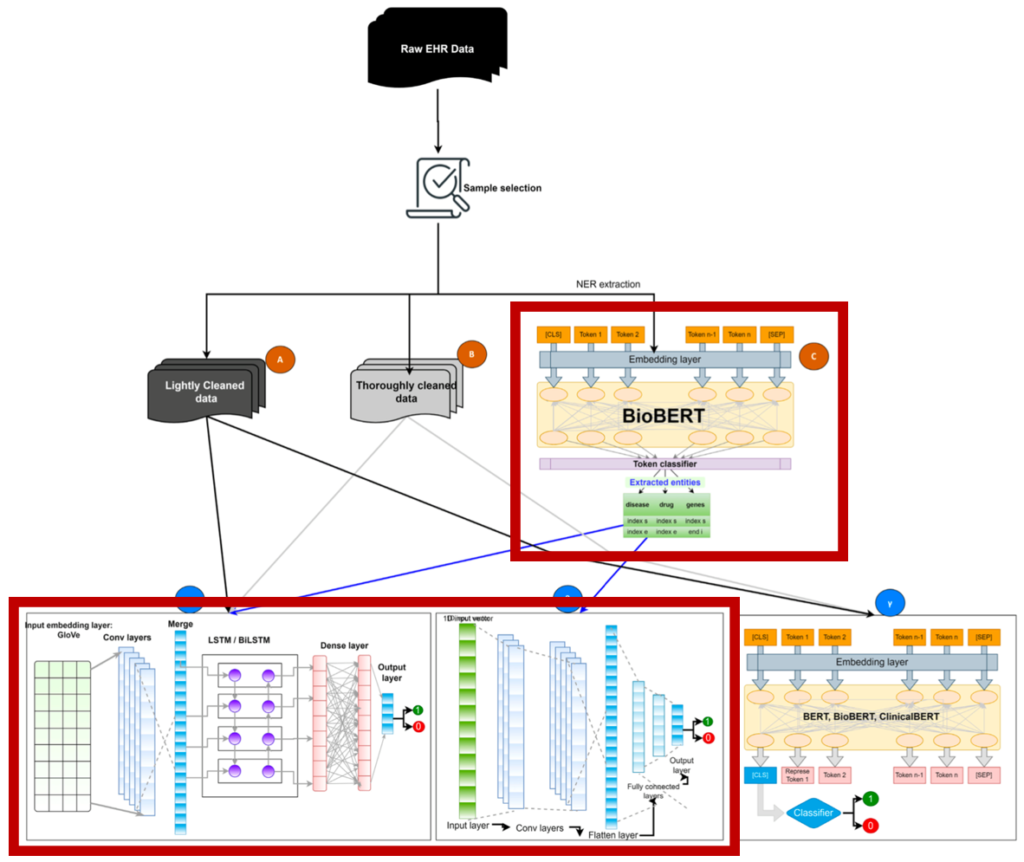
1. Clinical text data categorization and feature extraction using medical-fissure algorithm and neg-seq algorithm
- One of the pipelines for disease prediction uses only entities extracted from medical records.
- They cannot reflect relationships between entities and the entire text. Furthermore, there is a risk of losing meaningful information that can be obtained from other sentences.
2. Clinical text data categorization and feature extraction using medical-fissure algorithm and neg-seq algorithm
- Extending BERT for multi-type text classification and incorporating object-based medical knowledge graph.
- It is an independent framework for text and entity. Therefore, they cannot directly reflect the relationship between text and entities, and the model complexity is also high.
Proposed Method
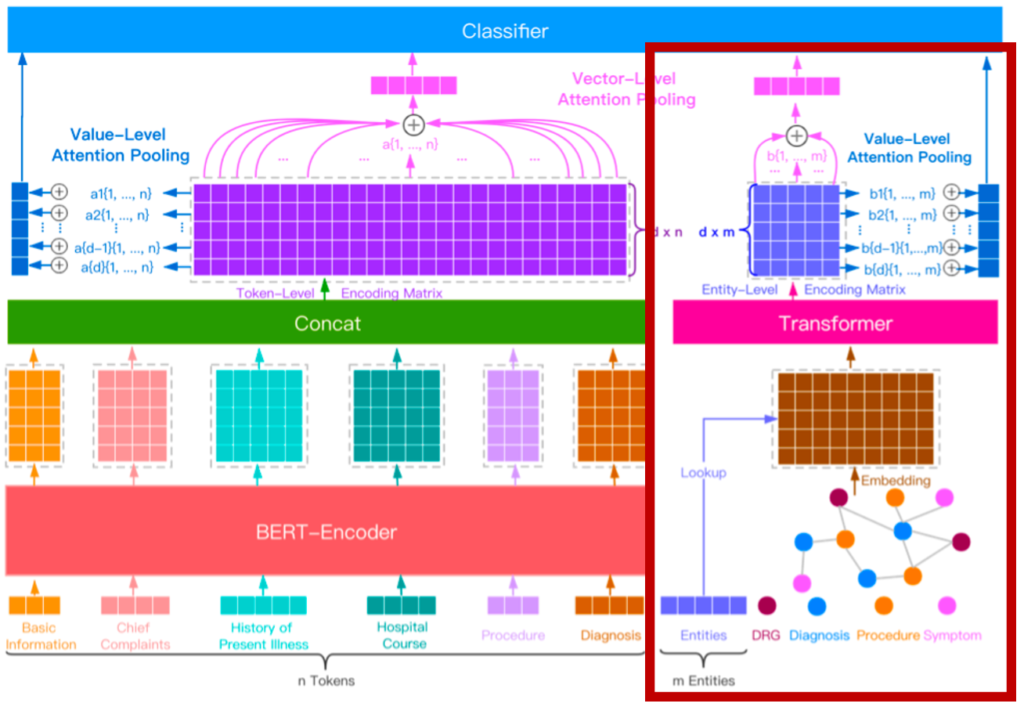
- In this study, we propose Entity-enhanced BERT (E-BERT), which utilizes the structural attributes of BERT for medical specialty prediction.
- E-BERT has an entity embedding layer and entity-aware attention to inject domain-specific knowledge and focus on relationships between medical-related entities within the sequences.
- E-BERT effectively incorporate domain-specific knowledge and other information, enabling the capture of contextual information in the text.